Section: New Results
Computational Physiology
Non-invasive personalisation of a cardiac electrophysiology model from body surface potential mapping
Participants : Sophie Giffard Roisin [Correspondent] , Maxime Sermesant, Nicholas Ayache, Hervé Delingette.
This work has been supported by the European Project FP7 under grant agreement VP2HF (no 611823) and the Marie Curie Actions European Industrial Doctorate CardioFunXion project (with Universitat Pompeu Fabra and Philips as partners).
Cardiac Modelling, Personalised Simulation, Inverse Problem of ECG, Electrical Simulation
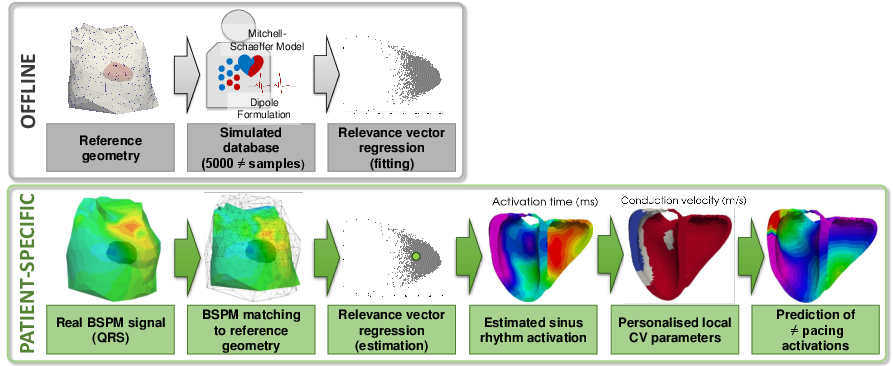
Within the VP2HF project, non-invasive cardiac electrical data has been acquired at St Thomas' Hospital, London. It consists in Body Surface Potential Mapping (BSPM), which are recordings of the electrical potential on several locations on the surface of the torso. In [37], we use non-invasive data (body surface potential mapping, BSPM) to personalise complex cardiac electrical activation patterns such as multiple onset activation locations. We have used a relevance vector regression (see Figure 18) and we have evaluated our method on clinical datasets.
Mulfidelity-CMA Personalisation Algorithm and Personalised 3D Modeling for Longitudinal Analysis
Participants : Roch Philippe Molléro [Correspondent] , Xavier Pennec, Hervé Delingette, Alan Garny, Nicholas Ayache, Maxime Sermesant.
This work has been partially funded by the EU FP7-funded project MD-Paedigree (Grant Agreement 600932) and contributes to the objectives of the ERC advanced grant MedYMA (2011-291080).
Cardiac Modelling, Personalised Simulation, Longitudinal Analysis, Parameter Estimation, Finite Element Mechanical Modelling
-
We extended the multiscale 0D/3D personalisation approach previously published to build a fast, flexible and computationally efficient multifidelity personalisation. This algorithm called Multifidelity-CMA can be used to personalise hundreds of cases per day without specific manual supervision, fine-tuning of the algorithm or precomputation. The method was published in a scientific journal [24].
-
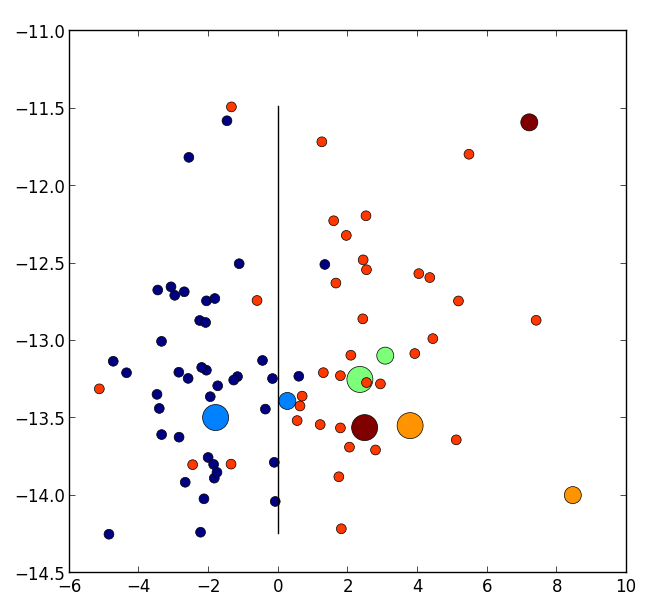
We built more than 140 personalised 3D simulations in the context of two longitudinal studies. We first used personalised parameters to model short-term transient effects in digestion ([33] and a poster presentation at FIMH Conference 2016), then to analyze long-term evolution of the cardiac function in cardiomyopathies ([42] and a poster presentation at MICCAI Conference 2017). In particular we showed that the use of priors reduces considerably the variance in the population of estimated parameters leading a better conditionning of parameter values whose variability in the population only reflects physiological properties of the cases. In particular we projected personalised parameters onto the axis of a classifier which discriminates between a cohort of healthy and diseased cases, and showed that the evolution of parameter values suggests an improvement of the cardiac function under therapy since the parameters of the follow-up acquisition are closer to the healthy side of the classifier (see Figure 19).
|